
CYTOSCAPE NETWORKS WITH BISOGENET SOFTWARE
This feature facilitates the integration of our software with other tools available in the Cytoscape platform.
In addition, an option is provided to allow other networks to be converted to BisoGenet. The client application includes network analysis tools and interactive network expansion capabilities. This feature could be instrumental to achieve a finer grain representation of the bioentities and their relationships. Two separate protein-protein interaction (PPI) networks of PP and HAD targets were constructed using the BisoGenet plug-in 23 and the intersection between the two PPIs identifed by Cytoscape 3.8. Methods: The data were imported into the STRING database to construct a protein-protein interaction network, and the network topology was analysed with the Bisogenet plug-in by Cytoscape 3.7.2. A feature of Bisogenet is the possibility to include coding relations to distinguish between genes and their products. Conclusion BisoGenet is able to build and visualize biological networks in a fast and user-friendly manner. The client tier is a Cytoscape plugin, which manages user input, communication with the Web Service, visualization and analysis of the resulting network. In the middle tier, a global network is created at server startup, representing the whole data on bioentities and their relationships retrieved from the database. In the data tier, an in-house database stores genomics information, protein-protein interactions, protein-DNA interactions, gene ontology and metabolic pathways. This is the output generated when Cytoscape is executed. Cytoscape recognizes a number of optional command line arguments, including run-time specification of network files, node and edge data files, and session files.
CYTOSCAPE NETWORKS WITH BISOGENET MANUAL
The BisoGenet network comprised of 1105 nodes. Command Line Arguments Cytoscape User Manual 3.10.0 documentation.
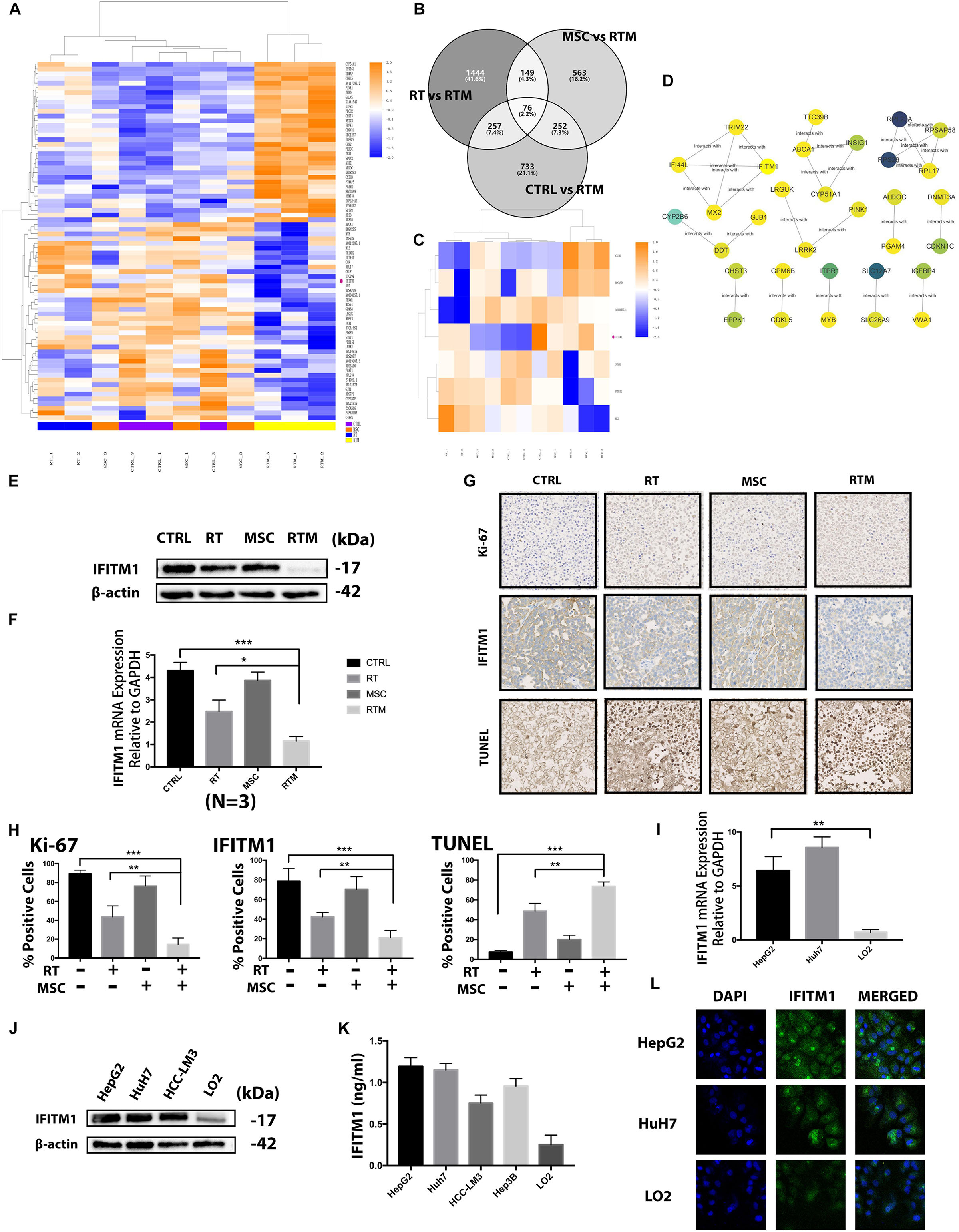
Results BisoGenet is a multi-tier application for visualization and analysis of biomolecular relationships. The overlapping DEGs were further mapped in gene-gene interaction network using BisoGenet in Cytoscape to analyze for significant hubs and modules. option is provided to allow other networks to be converted to BisoGenet. The median of three important attribute values, namely, degree, betweenness. The development of software tools capable of integrating data from different sources and to provide flexible methods to reconstruct, represent and analyze topological networks is an active field of research in bioinformatics. Then, the overlapping PPI network was obtained by using BisoGenet in Cytoscape. Several network tools have been implemented as Cytoscape plugins, including BisoGenet 3, MIMI 4 and APID2NET 5, which mainly focus on protein. Graph-based biological networks models capture the topology of the functional relationships between molecular entities such as gene, protein and small compounds and provide a suitable framework for integrating and analyzing omics-data. Abstract Abstract Background The increasing availability and diversity of omics data in the post-genomic era offers new perspectives in most areas of biomedical research. Imports interaction networks from public databases from a list of genes with their annotations and putative functions.


 0 kommentar(er)
0 kommentar(er)
